Vol. 101 No. 6 (2025)
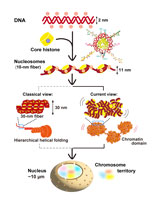
How 2 meters of human genome DNA are organized in the cell as chromatin has long been a fundamental question in cell biology. Negatively charged DNA is wrapped around positively charged core histones to form nucleosomes (left). For many decades, a string of nucleosomes (10-nm fiber) was believed to fold into regular 30-nm chromatin fibers and then into progressively thicker fibers in a hierarchical manner—forming the textbook model of chromatin structure (lower middle).
However, Maeshima et al. have challenged this classical paradigm (See the review article in this issue, pp. 339–356). Instead of static, regular fibers, chromatin in living higher eukaryotic cells forms a condensed domain organization that exhibits liquid-like properties (upper middle). These domains serve as functional units of chromatin that regulate access to DNA for key processes such as transcription and replication. On the right, these domains are shown to be assembled into a chromosome, which occupies a chromosome territory within the cell nucleus (shown in different colors).
The emerging “liquid-like domain” model highlights the viscoelastic nature of chromatin—locally fluid and dynamic to increase DNA accessibility, yet globally stable at the chromosome scale to preserve genome integrity. This model provides a conceptual framework for understanding how chromatin organization contributes to genome functions in space and time.
Yoshinori Ohsumi
Member of the Japan Academy
Table of Contents
-
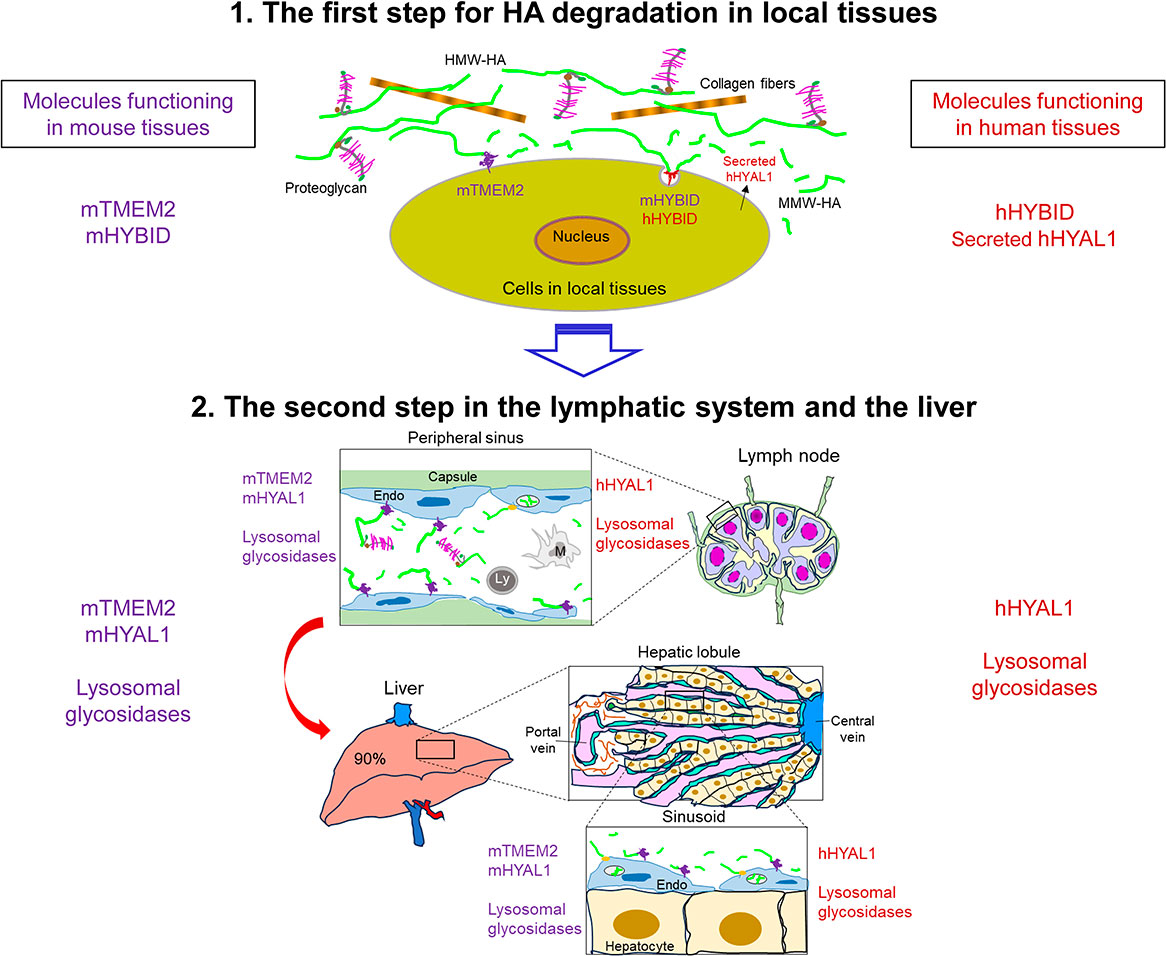
New molecules indispensable for hyaluronan degradation, HYBID (CEMIP/KIAA1199) and TMEM2 (CEMIP2): Differential roles in physiological and pathological non-neoplastic conditionsHiroyuki YOSHIDA, Shintaro INOUE and Yasunori OKADAVolume 101 Issue 6 Pages 317-338
-
The shifting paradigm of chromatin structure: from the 30-nm chromatin fiber to liquid-like organizationKazuhiro MAESHIMAVolume 101 Issue 6 Pages 339-356
-
f4-statistics-based ancestry profiling and convolutional neural network phenotyping shed new light on the structure of genetic and spike shape diversity in Aegilops tauschii Coss.Yoshihiro KOYAMA, Mizuki NASU and Yoshihiro MATSUOKAVolume 101 Issue 6 Pages 357-370
-
Correction to “Dynamics of the nucleoside diphosphate kinase protein DYNAMO2 correlates with the changes in the global GTP level during the cell cycle of Cyanidioschyzon merolae”Yuuta IMOTO, Yuichi ABE, Kanji OKUMOTO, Mio OHNUMA, Haruko KUROIWA, Tsuneyoshi KUROIWA and Yukio FUJIKI